Global Immunology and Immune Sequencing for Epidemic Response (GIISER) Sites in Africa

Overview
The emerging SARS-COV-2 variants of concern (VOC) demonstrate the urgent need for local scientific communities to have the capacity to rapidly detect and evaluate the immunological implications of new variants. The current VOC situation also provides the proving ground for such scientific collaborative infrastructure to inform and impact local and global equitable decision making around vaccines, herd immunity and therapeutics.
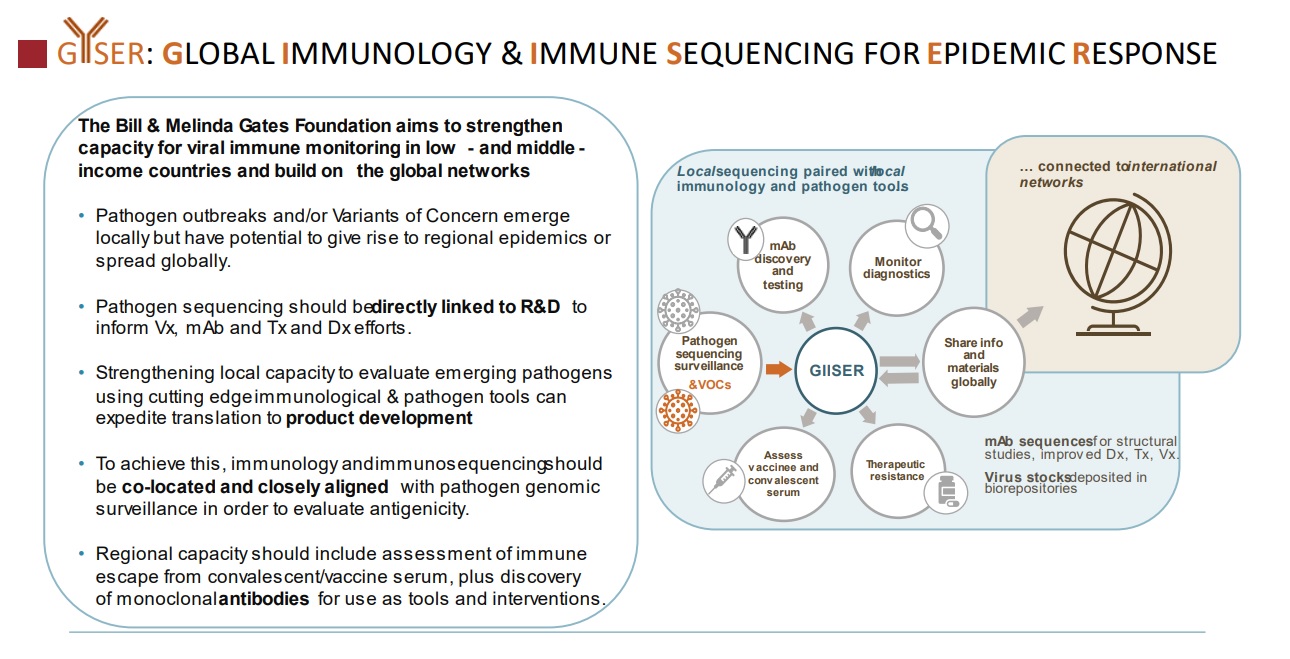
Global Immunology and Immune Sequencing for Epidemic Response (GIISER) is envisioned as a collection of geographically distinct sites that can pair their local pathogen sequencing surveillance and clinical epidemiology with local immunological understanding and tools. GIISER sites can connect via standard protocols, bioinformatics pipelines, data sharing guidelines and training opportunities to participate in coordinated international efforts to respond to emerging epidemic threats.
Importantly, the local immunological evaluation provided by GIISER extends to the rapid discovery of novel monoclonal antibodies that are critical tools for probing protein structure, detecting variant proteins, and identifying neutralizing epitopes for vaccine immunogen design. In addition, they may be developed as therapeutics for prevention or treatment of disease. Underlying all aspects of the work is the need for a global collaboration framework which includes rapid sharing of data and information with key international bodies (such as WHO, GISRS, and Africa CDC) and networks to inform an effective, equitable global pandemic response.
GIISER Project Goals
This specific effort aims to increase the in-country capacity for the rapid immunological evaluation of SARS-CoV-2 variants of concern on the African continent. It builds off of the African Pathogen Genomics Institute (PGI) sequencing capabilities (Tessema et al, Lancet Microbe Oct 2020), local human subjects research (clinical epidemiology), and adds in local virology and immunology expertise to allow in-country scientists to rapidly evaluate new variants for immune escape and cross protection. This data is essential for understanding the impact of new variants on vaccines, diagnostic assays, therapeutics and natural immunity.
The near-term goals of the GIISER program are 1. Systematic production of data quantifying immune escape and cross protection for variants of local concern/interest and 2. Identification of monoclonal antibodies that specifically recognize and potentially neutralize variants of local concern/interest isolated from convalescent individuals with sequence-confirmed SARS-CoV-2 infections.
A successful GIISER site will leverage existing pathogen sequencing surveillance infrastructure to rapidly identify emerging SARS-CoV-2 variants, obtain sera and blood cells from the appropriate infected individuals with appropriate informed consent, and perform detailed immunological characterization of sera/plasma (binding and neutralization assays), as well as single cell analytics for monoclonal antibody discovery. This information will be produced and communicated rapidly enough to inform public health decisions and policy makers within the local and international community. Many of these pieces already exist but are currently siloed. Some pieces need technological innovation to be rapid and effective.
Other pieces are nascent but training of local scientists by collaborators willing and able to share knowledge and tools will create the necessary capabilities.
A long-term goal is to establish a sustainable, local infrastructure that can be activated to respond to any new emerging pathogen locally and strengthen connections and collaborations between sites and with international decision-making bodies.
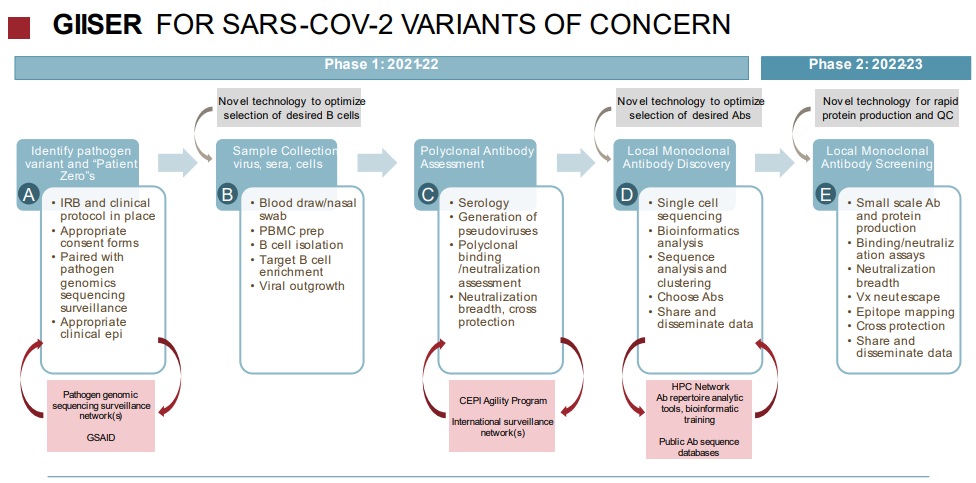
GIISER Scope of Work Outline (see Figure 1 below)
- Identify SARS-CoV-2 pathogen variants and appropriate subjects.
- Ethics approvals and clinical protocols in place with appropriate consent forms that allow for use of the donor sample for all downstream R&D which will support the connection of pathogen sequencing, longitudinal/contextual clinical data (including demographics and other relevant metadata) and subsequent immunological analysis. Build on capacity generated from prior initiatives (e.g., H3Africa human subjects research expertise and ethics frameworks)
- Pathogen sequencing and analytical infrastructure in place that meets international quality standards and shares sequence data publicly in a timely manner (via GISAID for example).
- Sample Collection, Virus Isolation and B cell sorting
- Laboratory capacity for virus sampling for sequencing and virus outgrowth (nasal swabs or other), including BSL3 capacity.
- Laboratory capacity for blood draws, including serum, plasma, PBMCs, B cell and/or plasmablast enrichment.
- Antigen specific B cell sorting via flow cytometry with tagged protein baits (generated locally or provided by collaborators).
- Assessment of natural- and vaccine-induced polyclonal antibody immunity to novel variants
- Quantitative assays measuring polyclonal antisera binding against both prior and novel VOC spike proteins (ELISA, MSD or other), calibrated against international standards or reference panels.
- Surrogate neutralization assays measuring inhibition of binding to ACE2 by polyclonal sera/plasma.
- Generation and use of VSV or lentivirus pseudoviruses containing novel spike protein sequences to quantitatively measure pseudovirus neutralization activity in sera/plasma, calibrated against international standards or reference panels.
- Implementation of live virus neutralization assays in BSL3 facilities to measure neutralization activity in polyclonal sera/plasma, calibrated against international standards or reference panels.
-
- Assessment of virus neutralizing activity in sera/plasma from prior infections with sequence- confirmed viral infections and vaccinated individuals to determine magnitude and breadth of natural and vaccine-induced cross neutralization.
- Coordinate with other GIISER sites, international networks (WHO, AfCDC, CEPI, etc.) to share high quality binding and neutralization results. Establish appropriate MTAs between sites to share materials.
- Monoclonal Antibody Discovery: sequencing and repertoire analysis
- Single cell immunoglobulin heavy and light chain sequencing from B cells enriched for binding to VoC spike proteins. This can be accomplished by any of a number of different technologies, including 10X, Seqwell, or others. Multiple specific baits and labels could be used for enrichment, targeting VOC specific mAbs (unique epitopes) or broadly neutralizing mAbs (conserved epitopes), or both.
- Proficiency in immunoglobulin sequence bioinformatics and clustering algorithms to assess somatic hypermutation, assign clonal lineages and evaluate Ab repertoire. Leverage opportunities such as those available via H3Bionet, Human Cell Atlas and GH-VAP to assist.
- Choose representative heavy + light chain pairs for subsequent expression and in vitro screening assays (See E).
- Engage with international antibody sequencing databases and consortia to share mAb repertoire information and sequences as appropriate, with future-use data sharing terms clearly outlined.
- Monoclonal Antibody Discovery: Screening and functional evaluation
- Small scale production of selected high quality mAbs from cDNA sequence.
- Systematic screening of mAbs for binding to a variety of spike proteins, including epitope mapping.
- Evaluation of neutralizing activity of selected mAbs against VOCs using the live virus and pseudovirus assays from step C above. Evaluate neutralization breadth and cross protection.
- Disseminate findings and engage with additional partners to share mAbs that have utility as either research tools or potential developability as therapeutics. Participate in international consortia such as COVIC when sufficient data supports such a move.
Evaluation Metrics and Criteria
GIISER sites will be evaluated based on their ability to link together pathogen sequencing, clinical epidemiology, immunology and monoclonal antibody discovery to rapidly produce high-quality, interpretable and informative data on neutralization breadth and cross protection for locally identified SARS-CoV-2 variants of concern. This includes, but is not limited to, outgrowth of variant SARS-CoV-2 virus in appropriate BLS3 containment labs for live virus neutralization assays. Examples of anticipated results: Cele et al Nature 2021, Moyo-Gwete et al NEJM 2021, Shen et al NEJM 2021 and Wibmer et al Nature Medicine 2021.
A key rationale for this effort is rapid data generation to inform decision making, thus the speed at which high quality data can be disseminated is an important consideration in the design and execution of each program. Within the first year of a two year investment, a well-functioning GIISER site will be communicating results from the first round of steps A-C within 1-2 months of identifying a variant of local concern, and steps D and E within an additional 2-3 months.
At the end of a two-year investment, we expect that each GIISER site will have a monoclonal antibody discovery pipeline that can feed lead candidates into product development. Of particular interest is to provide a pipeline for regionally distributed small footprint manufacturing which could further reduce the cycle time between discovery and delivery to LMICs
Sites will also be evaluated on their ability to leverage existing local expertise, collaborate and share data effectively with other GIISER sites and international networks (Figure 2 )
Requirements
A successful GIISER sites will have access to existing pathogen sequencing capacity and bioinformatics, access to existing cohorts or clinical infrastructure, access to existing BLS3 labs and trained virologists and immunologists with some expertise in protein reagent generation, mAb discovery and/or single cell analytics/bioinformatics. Not every site will have everything in place, but enthusiastic, creative and collaborative groups who want to get things done are encouraged to include specific requests for the support they need to get there (training, connections, technology, etc.).
What we are not looking to fund
We are not looking to fund pathogen genomics surveillance, expansion of surveillance sequencing capacity, or establishment of new clinical cohorts. We expect these to already be in place via other funding and for GIISER to build productive collaborations with these existing programs. Studies focused on T cell immunity and characterizing the overall immune response to SARS-CoV-2 are out of scope.
NOTE: This opportunity was open to an invited set of African investigators.